Overview of retrospective saddlepoint approximation (SPA) methods in GWAS
The retrospective saddlepoint approximation (SPA) methods are mainly designed to conduct genome-wide association studies (GWAS) in terms of both single-variant and set-based analysis.
Genome-wide association studies
All retrospective-SPA-based GWAS approaches share the same analysis framework including the following two steps
- Step 1: Fit a null model using trait, covariates, and GRM (if applied).
- Step 2: Conduct single-variant or set-based tests to identify marker or marker-set (e.g. gene) significantly associated with the trait of interest.
Genome-wide gene-environment interaction (GxE) studies
All retrospective-SPA-based GxE approaches share the same analysis framework including the following two steps:
- Step 1: Fit a genotype-independent (covariates-only) model using trait, covariates, and GRM (if applied).
- Step 2: Conduct single-variant or set-based tests to identify marker or marker-set (e.g. gene) significantly associated with the trait of interest.
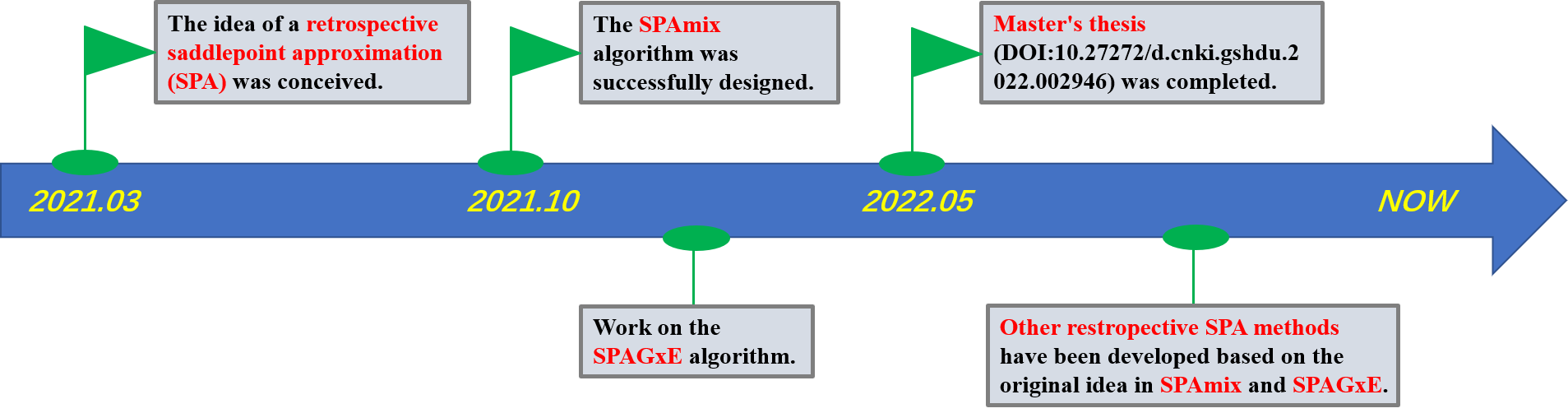
Timeline of the Retrospective SPA Project
March 2021
- Event: The idea of a retrospective saddlepoint approximation (SPA) was conceived.
October 2021
- Event: The SPAmix algorithm was successfully designed.
October 2021 - February 2022
- Event: Work on the SPAGxE algorithm was initiated.
May 2022
- Event: The master’s thesis (DOI:10.27272/d.cnki.gshdu.2022.002946), which initially introduced the concept of retrospective SPA, was completed.